Volume 8, Issue 3 (10-2020)
Jorjani Biomed J 2020, 8(3): 27-35 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Onsori H, Poladi D, Valizadeh M, Fathi A, Damandan M, Moradpour R. Molecular Identification of G6PD Chatham (1003 G>A) in North-West Iran. Jorjani Biomed J 2020; 8 (3) :27-35
URL: http://goums.ac.ir/jorjanijournal/article-1-743-en.html
URL: http://goums.ac.ir/jorjanijournal/article-1-743-en.html
Habib Onsori1 

 , Davood Poladi2
, Davood Poladi2 

 , Mehdi Valizadeh *3
, Mehdi Valizadeh *3 

 , Afshin Fathi4
, Afshin Fathi4 

 , Mahshid Damandan5
, Mahshid Damandan5 

 , Rouhallah Moradpour6
, Rouhallah Moradpour6 




 , Davood Poladi2
, Davood Poladi2 

 , Mehdi Valizadeh *3
, Mehdi Valizadeh *3 

 , Afshin Fathi4
, Afshin Fathi4 

 , Mahshid Damandan5
, Mahshid Damandan5 

 , Rouhallah Moradpour6
, Rouhallah Moradpour6 


1- Cellular and Molecular Biology Department, Marand Branch, Islamic Azad University, Marand, Iran
2- Department Genetics, Ahar Branch, Islamic Azad University, Ahar, Iran
3- Unit of Genomics Research, Digestive Diseases Research Center, Ardabil University of Medical Sciences, Ardabil, Iran ,mehdi_valizadeh65@yahoo.com
4- Pediatric Hematology & Oncology Department, Ardabil University of Medical Sciences, Ardabil, Iran
5- Center for Cell Pathology Research, Department of Life Science, Khazar University, Baku, Azerbaijan/Cellular and Molecular Research Center, School of Medicine, Ardabil University of Medical Sciences, Ardabil, Iran
6- Cellular and Molecular Research Center, School of Medicine, Ardabil University of Medical Sciences, Ardabil, Iran/Center for Cell Pathology Research, Department of Life Science, Khazar University, Baku, Azerbaijan
2- Department Genetics, Ahar Branch, Islamic Azad University, Ahar, Iran
3- Unit of Genomics Research, Digestive Diseases Research Center, Ardabil University of Medical Sciences, Ardabil, Iran ,
4- Pediatric Hematology & Oncology Department, Ardabil University of Medical Sciences, Ardabil, Iran
5- Center for Cell Pathology Research, Department of Life Science, Khazar University, Baku, Azerbaijan/Cellular and Molecular Research Center, School of Medicine, Ardabil University of Medical Sciences, Ardabil, Iran
6- Cellular and Molecular Research Center, School of Medicine, Ardabil University of Medical Sciences, Ardabil, Iran/Center for Cell Pathology Research, Department of Life Science, Khazar University, Baku, Azerbaijan
Keywords: Glucose 6-phosphate dehydrogenase (G6PD) , Chatham mutation , PCR-RFLP method , Sequencing
Full-Text [PDF 669 kb]
(3548 Downloads)
| Abstract (HTML) (9906 Views)
For precise evaluation of genetic changes of G6PD (Exon 9), sequencing method was randomly performed on a number of samples. For this purpose, the PCR product was sent to South Korea for sequencing with the help of Zhen Fanavaran Company and the results were analyzed by Chromas software.
.png)
Figure 1. Quality of extracted DNA on 1% agarose gel.
.png)
Figure 2. PCR product of the ninth exon fragment of G6PD on 2% agarose gel (L:50 bp Ladder, P: PCR Product)
Searching for Chatham mutation in PCR products:
After enzymatic digestion of PCR products by Bstsx1 enzyme, normal samples had one cleavage site for the enzyme and showed two fragments (130- 78 bp). On the other hand, samples with Chatham mutation had two cleavage sites for the enzyme and showed three fragments (30, 78, and 10 bp). Fragments 30 and 100 bp indicate the existence of mutation in the studied samples. Figure 3 display an example of enzymatic digestion by Bstx1. According to this figure, the plates 1, 3, and 4 have followed the 100 bp band pattern, indicating that the patients related to these plates are afflicted by Chatham mutation. On the other hand, the plates 2, 5, and 6 have followed the 130 bp band pattern which suggests that the patients related to these plates have no mutation (Fig 3).
.png)
Figure 3. Restriction digestion analysis of PCR products related to G6PD Chatham mutation with Bstx1. Lanes:2,5,6: No mutant. Lane 1,3,4: G6PD Chatham mutant.
Nucleotide Sequencing
To confirm and determine the location of Chatham mutation (G1003A) in Exon 9 of G6PD gene by primers (Table. 1). 2 mutated and one wild samples were sent to South Korea for sequencing with the help of Zhen Fanavaran Company. The results of sequencing were analyzed by Chromas lite version 2.33 software. The exact location of mutation has been presented in Figure 4, Figure 5, and Figure 6.
Table 1. Primers used in reactions

Figure 4. The mutated sample.
Figure 4 is related to nucleotide sequence of Exon 9 in Subject 19. This figure shows that GCC codon has turned to ACC at the position 1003. In fact, nucleotide transitional mutation of G to A has caused alanine 335 amino acid to be replaced by threonine (Ala 335 Thr).
.png)
Figure 6. The wild sample
Figure 6 is related to Exon 9 in Subject 39. According to this figure, GCC codon has not changed at the position 1003. This mean that this subject lacks G1003A mutation.
Result
This study was conducted on 90 individuals with deficient G6PD enzyme activity. Individuals’ ages ranged from one month to 21 years. G6PD Chatham mutation (1003 G>A) was observed in 10 (9 males and 1 females) cases, which represents the prevalence rate of 11.11% (Table 2).
Table 2. Gender differentiation of patients with Chatham mutation
Full-Text: (2972 Views)
Introduction
Glucose-6-Phosphate Dehydrogenase (G6PD; EC 1.1.1.49) deficiency was initially described in 1956. So far, researchers have conducted various studies on this subject (1). More than 400 G6PD variants have been identified up to date, of which 186 variants are linked to G6PD deficiency by decreasing the activity or stability of G6PD (2). The glucose-6-phosphate dehydrogenase (G6PD) is an enzyme which participates in first step of pentose phosphate pathway. G6PD converts glucose-6-phosphate to 6-phosphogluconolactone with the concomitant conversion of NADP+ to NADPH. The main function of NADPH is to keep adequate intracellular concentration of reduced forms of glutathione and other sulphydryl groups (2, 4).
NADPH enable cells to resist to oxidative damage by preserving and regenerating reduced forms of glutathione and promoting the stability of catalase.
Since the production of NADPH in erythrocytes is only rely on G6PD, so defense against oxidative damage is mainly depend on its activity (4).
The G6PD gene is located on the Xq28 chromosome consist of 13 exons and 12 introns with total length of 18.5 kb. Enzyme is active as a dimer or tetramer, each monomer is composed of 515 amino acid subunits with molecular weight around 59 kDa. G6PD deficiency is the most common inherited disorders in human being, affecting as many as 400 million people worldwide (3).
Although the majority of individuals with this disease may remain clinically asymptomatic, but some of the clinical manifestation including infection-, food- or drug- induced acute hemolytic anemia, newborns jaundice and chronic non-spherocytic hemolytic anemia (CNSHA) (5, 6).
In Iran, the overall prevalence of G6PD deficiency is about 10 to 14.9% (7, 8).
According to literatures, the most common mutations in Iran are Mediterranean mutation at nt 563 (C→T), Chatham mutation at nt 1003 (G→A) and Cosenza mutation at nt 1376 (G→C), respectively (8). The aim of the present study is to investigate the frequency rate of the Chatham mutation in north-west of Iran.
Materials and Methods
The study was conducted in Ardabil University of medical sciences, Ardabil Iran. In this study 90 unrelated patients with G6PD deficiency referred to hospitals in north-west of Iran (Ardabil, Tabriz and Urmia). Written consent was provided for each patient. Qualitative measurement of the enzyme activity was performed using Fluorescent Spot Test and Saba laboratory kit. The basis of this method is the catalytic activity of G6PDenzyme in conversion of G6PD Glucose 6-phosphate to 6-Phosphogloconate and simultaneous revival of NADP to NADPH2. The produced NADPH2 has fluorescent specifications under UV (365nm). The fluorescence intensity in the blood of healthy individuals is positive (strong) and in the blood of patients with G6PD deficiency is low or negative. The genomic DNA was extracted from Peripheral blood leukocytes by Rapid genomic DNA extraction (RGDE method) (9). After completing the extraction protocols, Quality of the extracted DNA products was evaluated in 1% agarose gel electrophoresis (Fig. 1). Afterward, samples from patients with G6PD deficiency were investigated by PCR-RFLP method. Conditions for PCR were as follows: 10 cycles for 30 sec at 95˚C, 1 min at 66˚C, followed by 1 min at 72˚C; and 20 cycles at 95˚C, 66˚C, and 72˚C for 1 min (10).
Each samples was visualized by using 1.5-2 % agarose gel electrophoresis to observe the nucleotide Fragment 208 of the ninth exon of G6PD (Fig. 2). Then, the proliferated product was digested by Bstx1 enzyme (Fermentas Co.) and subsequently electrophoresis was done on 3% agarose gel. Digestion reactions includes two steps of digestion and inactivation which were done at a temperature of 37˚C for 120 min and at 65˚C for 20 min.
Glucose-6-Phosphate Dehydrogenase (G6PD; EC 1.1.1.49) deficiency was initially described in 1956. So far, researchers have conducted various studies on this subject (1). More than 400 G6PD variants have been identified up to date, of which 186 variants are linked to G6PD deficiency by decreasing the activity or stability of G6PD (2). The glucose-6-phosphate dehydrogenase (G6PD) is an enzyme which participates in first step of pentose phosphate pathway. G6PD converts glucose-6-phosphate to 6-phosphogluconolactone with the concomitant conversion of NADP+ to NADPH. The main function of NADPH is to keep adequate intracellular concentration of reduced forms of glutathione and other sulphydryl groups (2, 4).
NADPH enable cells to resist to oxidative damage by preserving and regenerating reduced forms of glutathione and promoting the stability of catalase.
Since the production of NADPH in erythrocytes is only rely on G6PD, so defense against oxidative damage is mainly depend on its activity (4).
The G6PD gene is located on the Xq28 chromosome consist of 13 exons and 12 introns with total length of 18.5 kb. Enzyme is active as a dimer or tetramer, each monomer is composed of 515 amino acid subunits with molecular weight around 59 kDa. G6PD deficiency is the most common inherited disorders in human being, affecting as many as 400 million people worldwide (3).
Although the majority of individuals with this disease may remain clinically asymptomatic, but some of the clinical manifestation including infection-, food- or drug- induced acute hemolytic anemia, newborns jaundice and chronic non-spherocytic hemolytic anemia (CNSHA) (5, 6).
In Iran, the overall prevalence of G6PD deficiency is about 10 to 14.9% (7, 8).
According to literatures, the most common mutations in Iran are Mediterranean mutation at nt 563 (C→T), Chatham mutation at nt 1003 (G→A) and Cosenza mutation at nt 1376 (G→C), respectively (8). The aim of the present study is to investigate the frequency rate of the Chatham mutation in north-west of Iran.
Materials and Methods
The study was conducted in Ardabil University of medical sciences, Ardabil Iran. In this study 90 unrelated patients with G6PD deficiency referred to hospitals in north-west of Iran (Ardabil, Tabriz and Urmia). Written consent was provided for each patient. Qualitative measurement of the enzyme activity was performed using Fluorescent Spot Test and Saba laboratory kit. The basis of this method is the catalytic activity of G6PDenzyme in conversion of G6PD Glucose 6-phosphate to 6-Phosphogloconate and simultaneous revival of NADP to NADPH2. The produced NADPH2 has fluorescent specifications under UV (365nm). The fluorescence intensity in the blood of healthy individuals is positive (strong) and in the blood of patients with G6PD deficiency is low or negative. The genomic DNA was extracted from Peripheral blood leukocytes by Rapid genomic DNA extraction (RGDE method) (9). After completing the extraction protocols, Quality of the extracted DNA products was evaluated in 1% agarose gel electrophoresis (Fig. 1). Afterward, samples from patients with G6PD deficiency were investigated by PCR-RFLP method. Conditions for PCR were as follows: 10 cycles for 30 sec at 95˚C, 1 min at 66˚C, followed by 1 min at 72˚C; and 20 cycles at 95˚C, 66˚C, and 72˚C for 1 min (10).
Each samples was visualized by using 1.5-2 % agarose gel electrophoresis to observe the nucleotide Fragment 208 of the ninth exon of G6PD (Fig. 2). Then, the proliferated product was digested by Bstx1 enzyme (Fermentas Co.) and subsequently electrophoresis was done on 3% agarose gel. Digestion reactions includes two steps of digestion and inactivation which were done at a temperature of 37˚C for 120 min and at 65˚C for 20 min.
For precise evaluation of genetic changes of G6PD (Exon 9), sequencing method was randomly performed on a number of samples. For this purpose, the PCR product was sent to South Korea for sequencing with the help of Zhen Fanavaran Company and the results were analyzed by Chromas software.
.png)
Figure 1. Quality of extracted DNA on 1% agarose gel.
.png)
Figure 2. PCR product of the ninth exon fragment of G6PD on 2% agarose gel (L:50 bp Ladder, P: PCR Product)
Searching for Chatham mutation in PCR products:
After enzymatic digestion of PCR products by Bstsx1 enzyme, normal samples had one cleavage site for the enzyme and showed two fragments (130- 78 bp). On the other hand, samples with Chatham mutation had two cleavage sites for the enzyme and showed three fragments (30, 78, and 10 bp). Fragments 30 and 100 bp indicate the existence of mutation in the studied samples. Figure 3 display an example of enzymatic digestion by Bstx1. According to this figure, the plates 1, 3, and 4 have followed the 100 bp band pattern, indicating that the patients related to these plates are afflicted by Chatham mutation. On the other hand, the plates 2, 5, and 6 have followed the 130 bp band pattern which suggests that the patients related to these plates have no mutation (Fig 3).
.png)
Figure 3. Restriction digestion analysis of PCR products related to G6PD Chatham mutation with Bstx1. Lanes:2,5,6: No mutant. Lane 1,3,4: G6PD Chatham mutant.
Nucleotide Sequencing
To confirm and determine the location of Chatham mutation (G1003A) in Exon 9 of G6PD gene by primers (Table. 1). 2 mutated and one wild samples were sent to South Korea for sequencing with the help of Zhen Fanavaran Company. The results of sequencing were analyzed by Chromas lite version 2.33 software. The exact location of mutation has been presented in Figure 4, Figure 5, and Figure 6.
Table 1. Primers used in reactions
| Length of PCR product (bp) | Type of primer | Primer sequence | Exon |
| 208 |
Forward | 5ʹ CAA GGA GCC CAT TCT CTC CCT 3ʹ | 9 |
| Reverse | 3ʹ TTC TCC ACA TAG AGG ACG GCT GCC AAA GT 5ʹ |

Figure 4. The mutated sample.
Figure 4 is related to nucleotide sequence of Exon 9 in Subject 19. This figure shows that GCC codon has turned to ACC at the position 1003. In fact, nucleotide transitional mutation of G to A has caused alanine 335 amino acid to be replaced by threonine (Ala 335 Thr).

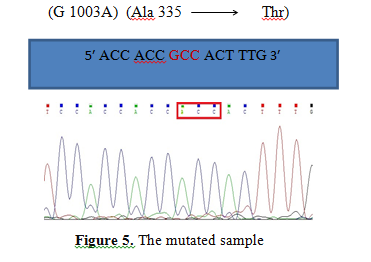
Figure 5. The mutated sample
Figure 5 is related to nucleotide sequence of Exon 9 in Subject 29. This figure shows that GCC codon has turned to ACC at the position 1003. In fact, nucleotide transitional mutation of G to A has caused alanine 335 amino acid to be replaced by threonine.
Figure 5 is related to nucleotide sequence of Exon 9 in Subject 29. This figure shows that GCC codon has turned to ACC at the position 1003. In fact, nucleotide transitional mutation of G to A has caused alanine 335 amino acid to be replaced by threonine.
.png)
Figure 6. The wild sample
Figure 6 is related to Exon 9 in Subject 39. According to this figure, GCC codon has not changed at the position 1003. This mean that this subject lacks G1003A mutation.
Result
This study was conducted on 90 individuals with deficient G6PD enzyme activity. Individuals’ ages ranged from one month to 21 years. G6PD Chatham mutation (1003 G>A) was observed in 10 (9 males and 1 females) cases, which represents the prevalence rate of 11.11% (Table 2).
Table 2. Gender differentiation of patients with Chatham mutation
| Female | Male | Total number of samples with Chatham mutation | Total number of patients |
| 1 (1.11%) |
9 (10%) |
10 (11.11%) |
90 |
It was also found that among these 10 patients with Chatham mutation, 5, 3, and 2 of them were from East Azerbaijan, West Azerbaijan, and Ardabil provinces (Table. 3).
Table 3. Geographic differentiation of patients with Chatham mutation
Table 3. Geographic differentiation of patients with Chatham mutation
| Ardabil | West Azerbaijan | East Azerbaijan | Total number of patients with Chatham mutation |
| 2 (2.2%) |
3 (3.3%) |
5 (5.5%) |
10 (11.11%) |
Discussion
The overall prevalence of G6PD deficiency among the Iranian population was 10%-14.9% (11).The present paper aimed to study the status of Chatham variant in northwest of Iran. Chatham variant of G6PD was firstly observed in a Hindi boy living in London with jaundice symptoms and then occurred in different populations. The highest and the lowest frequency of G6PD Chatham has been reported from north of Iran (27%) and Brazil (0.66%), respectively (12). There are multiple studies conducted on Chatham mutation around world on different ethnic groups. Altay (13) reported the frequency of Chatham mutation to be 2% in 50 patients with G6PD deficiency in Turkey. Al-Alawi (14) found a frequency of 87% for Chatham mutation among 580 men of the Kurds in northern Iraq. In a molecular study conducted by Carmencita (15) in Philippines, frequency of Chatham mutation was reported to be 9.4%. Tantular (16) carried out a molecular study on 99 patients with G6PD deficiency in 8 different ethnic groups in East Indonesia and found than 17.7% of them were afflicted with type of mutation.
Chatham mutation (1003 G>A) is caused by single-base mutation in Exon 9 of G6PD gene. The proliferated fragment in a healthy person has one recognition site for Bstx1 enzyme cutting, so it generates two fragments of 130 and 78 bp. In a patient, Chatham mutation creates a new cutting site in Fragment 130 bp and, as a result, three fragments of 30, 78, and 100 bp will be generated.
According to the results of the present study, it was found that 10 subjects (11.11%) had a Chatham mutation. Prevalence of Chatham mutation in this study (including 3 province) is less than its prevalence in Mazandaran (27%) and Khorasan (12%) but more than Fars (8.82%), Hormozgan (8.2%), Kermanshah (7.3%), Sistan and Baluchistan (2.17%), Khuzestan (8.66%), Isfahan (8.06%), and Yazd (3.6%) provinces (Table.4). This finding may imply the fact that different ethnic groups living in Iran may receive mutation from various sources.
Conclusion
The present paper aimed to study the status of Chatham variant in northwest of Iran.
The results showed that 10 (11.11%) out of 90 patients had a Chatham mutation. It should be noted that of the 90 patients participated in this study, 81 patients (92.23%) were male and others were female (7.77%). Also, among the 10 patients with Chatham mutation, 9 ones were male and only one was female. This data indicates the high prevalence of G6PD deficiency in males and its sex-linked inheritance pattern.
Prevalence of Chatham mutation in the three studied provinces (West Azerbaijan, East Azerbaijan, and Ardabil) of Iran is less than its prevalence in Mazanadaran (27%) and Khorasan (12%) but more than Fars (8.82%), Hormozgan (8.2%), Kermanshah (7.3%), Sistan and Baluchistan (2.17%), Khuzestan (8.66%), Isfahan (8.06%), and Yazd (3.6%) provinces. These results show that the prevalence of Chatham mutation in northwest of Iran is much more than southern regions and less than northern parts of Iran [Table.4]. Chatham mutation is the second most common mutation (after Mediterranean mutation) in some provinces of Iran (18). Molecular analysis of G6PD deficiency in this study conducted in North-West of Iran revealed that G6PD Chatham (1003 G>A) mutation is the second common mutation, after Mediterranean (563C>T) (8), in the population of the North-West of Iran. Further studies are recommended to identify the mutation type of other varieties.
Table 4. Prevalence of Chatham mutation in northern and southern provinces of Iran
The overall prevalence of G6PD deficiency among the Iranian population was 10%-14.9% (11).The present paper aimed to study the status of Chatham variant in northwest of Iran. Chatham variant of G6PD was firstly observed in a Hindi boy living in London with jaundice symptoms and then occurred in different populations. The highest and the lowest frequency of G6PD Chatham has been reported from north of Iran (27%) and Brazil (0.66%), respectively (12). There are multiple studies conducted on Chatham mutation around world on different ethnic groups. Altay (13) reported the frequency of Chatham mutation to be 2% in 50 patients with G6PD deficiency in Turkey. Al-Alawi (14) found a frequency of 87% for Chatham mutation among 580 men of the Kurds in northern Iraq. In a molecular study conducted by Carmencita (15) in Philippines, frequency of Chatham mutation was reported to be 9.4%. Tantular (16) carried out a molecular study on 99 patients with G6PD deficiency in 8 different ethnic groups in East Indonesia and found than 17.7% of them were afflicted with type of mutation.
Chatham mutation (1003 G>A) is caused by single-base mutation in Exon 9 of G6PD gene. The proliferated fragment in a healthy person has one recognition site for Bstx1 enzyme cutting, so it generates two fragments of 130 and 78 bp. In a patient, Chatham mutation creates a new cutting site in Fragment 130 bp and, as a result, three fragments of 30, 78, and 100 bp will be generated.
According to the results of the present study, it was found that 10 subjects (11.11%) had a Chatham mutation. Prevalence of Chatham mutation in this study (including 3 province) is less than its prevalence in Mazandaran (27%) and Khorasan (12%) but more than Fars (8.82%), Hormozgan (8.2%), Kermanshah (7.3%), Sistan and Baluchistan (2.17%), Khuzestan (8.66%), Isfahan (8.06%), and Yazd (3.6%) provinces (Table.4). This finding may imply the fact that different ethnic groups living in Iran may receive mutation from various sources.
Conclusion
The present paper aimed to study the status of Chatham variant in northwest of Iran.
The results showed that 10 (11.11%) out of 90 patients had a Chatham mutation. It should be noted that of the 90 patients participated in this study, 81 patients (92.23%) were male and others were female (7.77%). Also, among the 10 patients with Chatham mutation, 9 ones were male and only one was female. This data indicates the high prevalence of G6PD deficiency in males and its sex-linked inheritance pattern.
Prevalence of Chatham mutation in the three studied provinces (West Azerbaijan, East Azerbaijan, and Ardabil) of Iran is less than its prevalence in Mazanadaran (27%) and Khorasan (12%) but more than Fars (8.82%), Hormozgan (8.2%), Kermanshah (7.3%), Sistan and Baluchistan (2.17%), Khuzestan (8.66%), Isfahan (8.06%), and Yazd (3.6%) provinces. These results show that the prevalence of Chatham mutation in northwest of Iran is much more than southern regions and less than northern parts of Iran [Table.4]. Chatham mutation is the second most common mutation (after Mediterranean mutation) in some provinces of Iran (18). Molecular analysis of G6PD deficiency in this study conducted in North-West of Iran revealed that G6PD Chatham (1003 G>A) mutation is the second common mutation, after Mediterranean (563C>T) (8), in the population of the North-West of Iran. Further studies are recommended to identify the mutation type of other varieties.
Table 4. Prevalence of Chatham mutation in northern and southern provinces of Iran
| Ref | Prevalence | Province | Areas |
| (17) | 27% | Mazandaran | Northern provinces |
| (12) | 26.7% | Golestan | |
| (19) | 9.7% | Guilan | |
| (21) | 12% | Khorasan | |
| 11.11% |
Ardabil Tabriz Urmia |
Northwest provinces |
|
| (18,23) | 2.1% | Sistan and Baluchistan | Southern provinces |
| (20) | 8% | Hormozgan | |
| (22,24) | 8.66% | Khuzestan |
Acknowledgements
I would like to appreciate the respected authorities of the Hematology Laboratory of Koodakan-E-Tabriz (children hospital of Tabriz), Bu Ali Ardabil and shahid Motahari hospital of Urmia to give samples and also I would like to appreciate authorities of the Islamic Azad University of Marand to allow for the use of equipment and facilities of Genetics Laboratory.
I would like to appreciate the respected authorities of the Hematology Laboratory of Koodakan-E-Tabriz (children hospital of Tabriz), Bu Ali Ardabil and shahid Motahari hospital of Urmia to give samples and also I would like to appreciate authorities of the Islamic Azad University of Marand to allow for the use of equipment and facilities of Genetics Laboratory.
Type of Article: Original article |
Subject:
Molecular Sciences
Received: 2020/07/16 | Accepted: 2020/08/3 | Published: 2020/10/1
Received: 2020/07/16 | Accepted: 2020/08/3 | Published: 2020/10/1
References
1. Khodashenas E, Kalani-Moghaddam F, Araghi Z, Khodaparast, M, Yazdani, Z. Glucose-6-Phosphate Dehydrogenase Deficiency and Neonatal Hyperbilirubinemia. Iranian Journal of Neonatology 2015; 6(3): 28-31.doi: 10.22038/ijn.2015.4897. [Google Scholar]
2. Lee J, Kim TI, Kang J, et al. Prevalence of glucose-6-phosphate dehydrogenase (G6PD) deficiency among malaria patients in Upper Myanmar. BMC Infect Dis. 2018; 18, 131.
https://doi.org/10.1186/s12879-018-3031-y [view at publisher] [DOI] [Google Scholar]
3. Lo E, Zhong, D, Raya B. et al. Prevalence and distribution of G6PD deficiency: implication for the use of primaquine in malaria treatment in Ethiopia. Malar J. 2019; 18, 340.
https://doi.org/10.1186/s12936-019-2981-x [view at publisher] [DOI] [Google Scholar]
4. Mehta A, Mason P, and Valliamy TJ. Glocuse-6-phosphate dehydrogenase deficiency. Bailliers Clinical Hematology. 2002;13: 21-38. [view at publisher] [DOI] [Google Scholar]
5. Beutler E. Molecular heterogeneity of glucose-6-phasphate dehydrogenase. Blood. 1989; 74: 2550. [DOI] [Google Scholar]
6. Haung Ch.-Sh. Neonatal jaundice and molecular mutation in G6PD deficient newborn infants. American Journal of Hematology.1995; 51: 19-25.
https://doi.org/10.1002/(SICI)1096-8652(199601)51:1<19::AID-AJH4>3.0.CO;2-A [view at publisher] [DOI] [Google Scholar]
7. Noori-Daloii MR. A comprehensive study on the major mutations in G6PD deficient polymorphic variants identified in the coastal provinces of Caspian Sea in the north of Iran. Clin Biochem. 2007; 40: 699-704. [view at publisher] [DOI] [Google Scholar]
8. Valizadeh M, Onsori H, Fathi A, Bonyadi MJ, Rezamand A, Amani F. Molecular Identification of Mediterranean Mutation in Patients with Deficiency of Glucose-6-Phosphate Dehydrogenase (G6pd) in North West of Iran. J Phys Pharm Adv. 2014; 4(7): 389-395. [DOI] [Google Scholar]
9. Saremi Mohammad Ali et al. Rapid genomic DNA extraction (RGDE). Protocol Online PID. 2010; 4791, http://www.protocol-online.org [Google Scholar]
10. Noori Daloii MR, Najafi L, Mohammad Ganji S, Hajebrahimi Z, Sanati MH. Molecular identification of mutations in G6PD gene in patients with favism in Iran. J Physol Biochem.2004; 60(4); 273-7. [view at publisher] [DOI] [Google Scholar]
11. WHO Working Group. World map of G6PD deficiency. Bull WHO.1989; 67: 601-611. [Google Scholar]
12. Moosazadeh M, Nekoei-Moghadam M, Aliramzany M, Amiresmaili M. Identification of Mutation of Glucose-6-Phosphate Dehydrogenase (G6PD) in Iran: Meta- analysis Study. Iranian J Publ Health. 2013;42( 9): 1007-1015. [view at publisher] [Google Scholar]
13. Altay C, Gümrük F. Red cell glucose-6-phosphate dehydrogenase deficiency in Turkey. Turkish Journal of Haematology : Official Journal of Turkish Society of Haematology. 2008 Mar;25(1):1-7. [view at publisher] [Google Scholar]
14. Al-Allawi N, Eissa AA, Jubrael JM, Jamal SA, Hamamy H. Prevalence and molecular characterization of Glucose-6-Phosphate dehydrogenase deficient variants among the Kurdish population of Northern Iraq. BMC Blood Disord. 2010 Jul 5;10:6. doi: 10.1186/1471-2326-10-6. PMID: 20602793; PMCID: PMC2913952. [view at publisher] [DOI] [Google Scholar]
15. Carmencita P, Cutiongco E M, Silao L. Characterization of Mutations and Polymorphisms in the G6PD Gene among Filipino Newborns with G6PD Deficiency Acta medica Philippina.2011 45(4):53-57. [Google Scholar]
16. Tantular IS, Matsuoka H, Kasahara Y, Pusarawati S, Kanbe T, Tuda JS, Kido Y, Dachlan YP, Kawamoto F. Incidence and mutation analysis of glucose-6-phosphate dehydrogenase deficiency in eastern Indonesian populations. Acta Med Okayama. 2010 Dec;64(6):367-73. doi: 10.18926/AMO/41322. PMID: 21173806. [Google Scholar]
17. Mesbah Namin SA, Sanati MH, Mowjoodi A, Mason PJ, Vulliamy TJ, Noori Daloii MR. Three major glucose-6-phosphate dehydrogenase-deficient polymorphic variants identified in Mazandaran state of Iran. Br J Haematol. 2002; 117(3):763-4. [view at publisher] [DOI] [Google Scholar]
18. Mortazavi Y, Mirzamohammadi F, Teremahi Ardestani M, Mirimoghadam E, Vulliamy TJ. Glucose 6-phosphate dehydrogenase deficiency in Tehran, Zanjan and Sistan- Balouchestan provinces: prevalence and frequency of Mediterranean variant of G6PD. Iran J Biotechnol. 2010; 8(4):229-233. [view at publisher] [Google Scholar]
19. Noori-Daloii MR, Hajebrahimi Z, Najafi L. Molecular identification of the most prevalent mutation of glucose-6-phosphate dehydrogenase gene in deficient patients in Gilan province. J Sci IR Iran. 2003; 14(4): 327-31. [Google Scholar]
20. Noori-Daloii MR, Hejazi, SH, Yousefi A, Mohammad ganji S, Soltani S, Javadi KR, et al. Identification of Mutations in G6PD Gene in Patients in Hormozgan Province of Iran. J Sci I R Iran. 2006; 17(4): 313-316. [view at publisher] [Google Scholar]
21. Noori-Daloii MR, Soltanian S, Mohammad Gangi SH, Yousefi A, Hejazi S, Banihashem A, et al. Molecular Identification of the Most Prevalent Mutations of Glucose-6-Phosphate Dehydrogenase (G6PD) Gene in deficient Patients in Khorasan Province of Iran. J Sci I R Iran. 2006; 17(2):103-106. [Google Scholar]
22. Gandomani MG, Khatami SR, Kazeminezhad SR, Daneshmand S, Mashayekhi A. Molecular Identification of G6PD Chatham (G1003A) in Khuzestan Province of Iran. Journal of Genetics. 2011; 90(1):143-145. [DOI] [Google Scholar]
23. Noori-Daloii MR, Yousefi A, Mohammad Ganji S, Hejazi S, Soltanian S, Sanei Moghadam E, Bozorgzade P, Sanati MH. Molecular identification of the most prevalent mutations of g6pd gene in deficient patients in Sistan and Baluchestan province of Iran. J Sci IR Iran. 2005; 16(4): 321-2. [view at publisher] [Google Scholar]
24. Kazemi Nezhad SR, Mashayekhi A, Khatami SR, Daneshmand S, Fahmi F, Ghaderigandmani M, et al. Prevalence and Molecular Identification of Mediterranean Glucose-6-Phosphate Dehydrogenase Deficiency in Khuzestan Province, Iran. Iranian J Publ Health. 2009; 38(3):127.] [Google Scholar]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |



